Chromatin Modifications and Epigenetics in Human Disease
Epigenetics (表观遗传学)
- Epigenetics: the study of heritable changes in
gene expressionthat do not involve changes to the underlying DNA sequence
How is epigenetic information encoded?
- DNA methylation
- Location of nucleosomes
- specific histone
- histone modifications (on histone tails)
DNA regulatory elements Recap:
- Promotors: Control direction and amount of transcription
- Enhancers: Increase transcription
- Silencers: Set the boundaries of enhancer activity
DNA methylation
Facts:
- Cytosine (C) bases (胞嘧啶碱基, a pyrimidine base found in DNA and RNA) can be methylated (甲基化, the addition of a methyl group to a molecule)
- Mostly occurs at
CpGsites
(CpG site: a pair of cytosine and guanine nucleotides that are located next to eachother)
Regions with
high CpG densityare calledCpG islandsIs performed by DNA methyltransferases (DNMTs, DNA甲基转移酶)
Causes
chromatin condensationandgene repressionPromotoroffen asocciated with CpG islandsCan prevents
transcription factorbinding
DNA methylation patterns:
- Methylation leads to chromatin condensation and gene repression
- Differential methylation in imprinting control regions leads to genomic imprinting
imprinting (基因印记,遗传印记): a phenomenon in which the expression of a gene depends on whether it was inherited from the mother or the father
- Changes in methylation patterns during development
- Regulated by CTCF binding - methylation prevents CTCF binding
Wild type: the normal DNA sequence
Imprinting disorders:
- Silver-Russell syndrome: hypomethylation of ICR1, reduced expression of IGF2
- Beckwith-Wiedemann syndrome: hypermethylation of ICR1, increased expression of IGF2
- Angelman syndrome: loss of maternal UBE3A expression
- Prader-Willi syndrome: loss of paternal expression of NDN, SNORD11
Histone modifications
- Histone types: H2A, H2B, H3, H4
- How to read modification names: H3K4me3
- H3: histone type
- K4: amino acide modified
- me3: type of modification
Common Marks
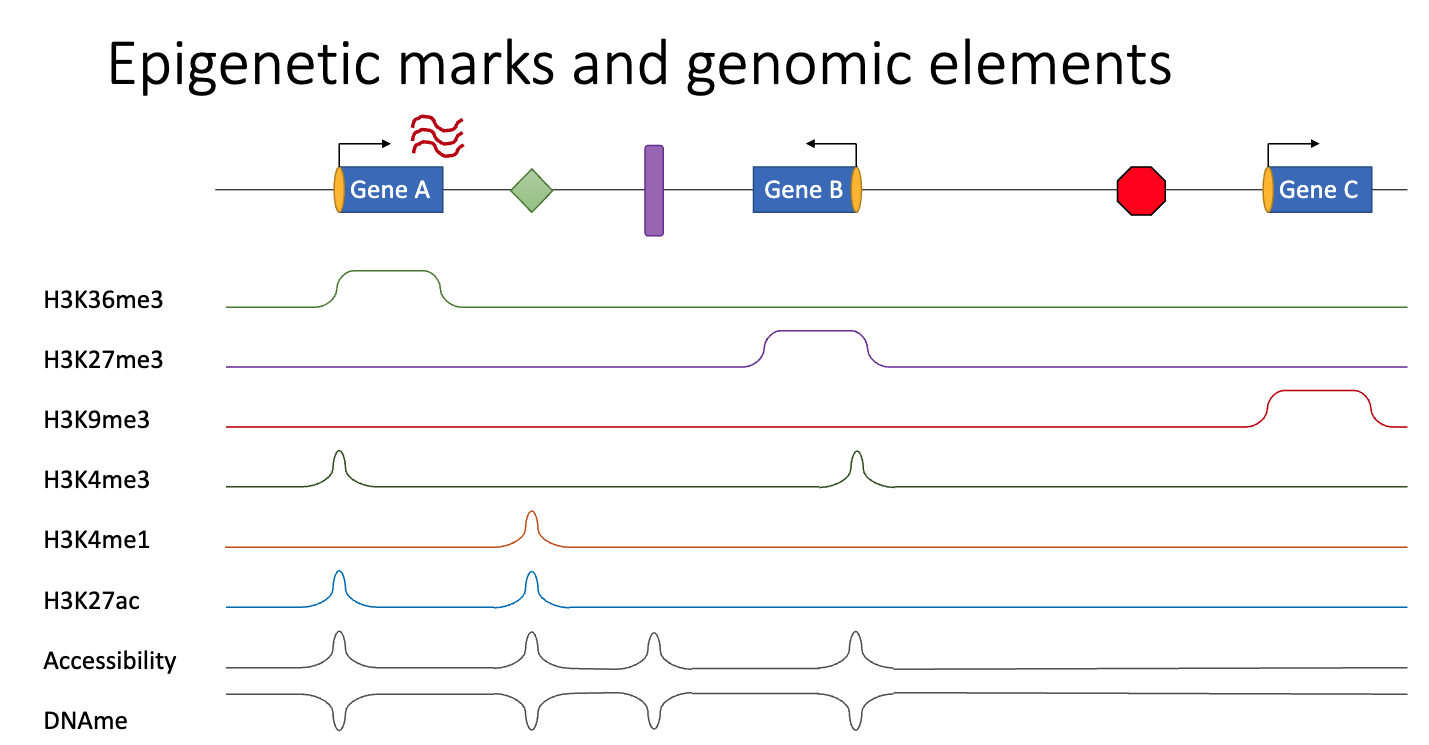
Beagrie, R., & Stolper, R. (2023, Oct 17). CM1.4. Chromatin Modifications and Epigenetic Factors [Epigenetic Marks and Genomic Elements]. University of Oxford.
- H3K36me3: Bodies of actively transcribed genes
- H3K27me3: Promoter and body of transiently/developmentally (Polycomb) repressed genes
- H3K9me3: Promoter and body of strongly repressed genes. Also repeat elements
- H3K4me3: Promoter of actively transcribed and (some) Polycomb repressed genes
- H3K4me1: Enhancers
- H3K27ac: Active enhancers and promoter of actively transcribed genes
- Accessibility: Enhancers, active promoters, Polycomb repressed promoters, insulators
- DNAme: CpGs at Enhancers, active promoters, Polycomb repressed promoters & insulators are demethylated
Chromatin Proteins
- Types of chromatin proteins:
- Readers: recognize and bind to specific histone modifications
- Writers: add histone modifications
- Erasers: remove histone modifications
- Remodelers: change the position of nucleosomes
Writers can be readers at some mark
Erasers can be readers at some mark
Mutation in theses proteins can lead to disease
Epigenetic diseases:
- Rubinstein-Taybi syndrome: mutation in
CBPorP300, intellectual disability, facial abnormalities, broad thumbs and toes