Principals Underlying Basic Molecular Genetic Techniques
Key Technologies to Study and Analyze Genes
DNA Cloning
- purify and amplify DNA fragments of interest
- recombinant DNA technology
- Recombinant DNA: DNA that has been created artificially by combining DNA from different sources
- DNA fragments lack of capability of initiating replication
- Replicon: Replication origin in a molecules
- Human DNA fragments need a suitable replicon to replicate within a bacterial cell.
- The result is the formation of a recombinant DNA.
Insertion of DNA Fragments into Cloning Vectors
Overview
- Isolated human DNA is long and needs to be fragmented for cloning.
- Cloning vectors can only accept relatively small DNA molecules.
- Fragmentation by physical shearing is not sufficient as it produces long fragments with heterogeneous ends.
- To facilitate ligation, small DNA fragments with uniform ends are required.
Tools Required
- Restriction Enzymes: Bacterial enzymes that recognize and cleave specific DNA sequences.
- DNA Ligases: Enzymes that join DNA fragments together.
Restriction Endonucleases
- They recognize specific short sequence elements within double-stranded DNA.
- Cleavage occurs on both strands within or close to these sequences.
- Bacteria produce restriction endonucleases with different sequence specificities.
Restriction Sites
- Often short palindromic sequences.
- Read the same in the 5ʹ to 3ʹ direction on both DNA strands.
- Cleavage may produce overhanging 5ʹ or 3ʹ ends (
sticky ends) or blunt ends.
Cloning Vectors
- DNA fragments with sticky or blunt ends can be inserted into vectors.
- Vectors are engineered to contain unique restriction sites for specific enzymes.
Plasmid Vectors
-
Double stranded, circular DNA molecules
-
E. coli plasmids are widely used as cloning vectors
-
Components:
- Origin of replication: allows the plasmid to replicate in E. coli
- Selectable marker: gene that confers antibiotic resistance
- Multiple cloning site: region with multiple unique restriction sites for insertion of DNA fragments
DNA Library
-
Genomic library:
- DNA library: DNA molecules are cloned into a vector molecule.
- Genomic library: a collection of
DNAfragments that represent theentire genomeof an organism.
-
Complementary DNA (cDNA) library:
- cDNA library: represents the mRNA expressed in a cell type.
- only
expressed genesare represented.
DNA Cloning Limitations
-
Laborious and time-consuming
-
Not suitable for parallel amplification of many DNA fragments from diferent sources
PCR (Polymerase Chain Reaction)
PCR is a technique that allows the amplification of a specific DNA fragment with gene-specific primers.
It is a cell-free method
Pros:
- Fast (minimum 1 hour)
- Parallel amplification of many DNA fragments from different sources
- Highly sensitive (can detect a single DNA molecule)
The Polymerase Chain Reaction (PCR) is a technique used in molecular biology to amplify a segment of DNA. Below is the summary in key steps:
PCR Steps
Enzoklop, CC BY-SA 4.0 https://creativecommons.org/licenses/by-sa/4.0, via Wikimedia Commons
-
Preparation: Gather necessary components:
- DNA template with the target sequence.
- Heat-stable DNA polymerase enzyme.
- Two PCR primers for the start and end of the target sequence.
- Deoxynucleotide triphosphates (dNTPs), the building blocks for new DNA.
- Reaction buffer with Magnesium ions for enzyme activity.
- A thermal cycler for temperature control.
-
Denaturation: Heat the mixture to make the DNA single-stranded.
-
Annealing: Cool down so primers can bind to their complementary DNA sequences.
-
Extension: Raise temperature for DNA polymerase to synthesize new DNA strands.
-
Amplification: Repeat the cycle multiple times, doubling the DNA amount each cycle.
-
Finalization: End with billions of copies of the specific DNA segment.
This process allows for the DNA to be sufficiently amplified for further analysis.
PCR Cycle
- Denaturation
- Annealing
- Extension
This Cycle is repeated 25-35 times
PCR Primers
Designing PCR Primers
-
Specificity:
- Primer sequence: Primers should only bind to the target sequence.ce:
- Length: 17-28bp optimal.
- Nature of the template:
- Melting temperature (Tm):
- Optimal Tm: 52-65°C
- Influenced by GC content, length, and concentration
- Anneling temperature: 5°C below Tm
- Working Approximation: Tm = 4(G+C) + 2(A+T)
- GC content: 45-55% optimal
- Melting temperature (Tm):
-
Secondary structures and primers homology
- Avoid regions of self-homology (within the sequence)
Self-homology: the presence of complementary regions within a single nucleic acid molecule.
- Avoid inter-primer homology that could lead to primer dimers (between two primers)
inter-primer homology: the presence of complementary regions between two different primers.
-
Amplicon size
- Avoid large amplicon size
-
Repeats and Runs
- Long runs of single base repeats larger than 4bp should be avoided
- Long runs of di-nucleotide repeats should be avoided
Gene Expression Analysis Techniques
Endpoint of PCR
Measures the amount of DNA at the end of the reaction, rather than during each cycle. It is semi-quantitive which means that it can be used to compare the amount of DNA in different samples, but it cannot be used to determine the exact amount of DNA in a sample.
- Easy and Cheap
- Not very accurate
Angarose gel electrophoresis is used to separate DNA fragments by size. The DNA is stained with a dye that fluoresces under UV light. The DNA is visualized as bands on the gel.
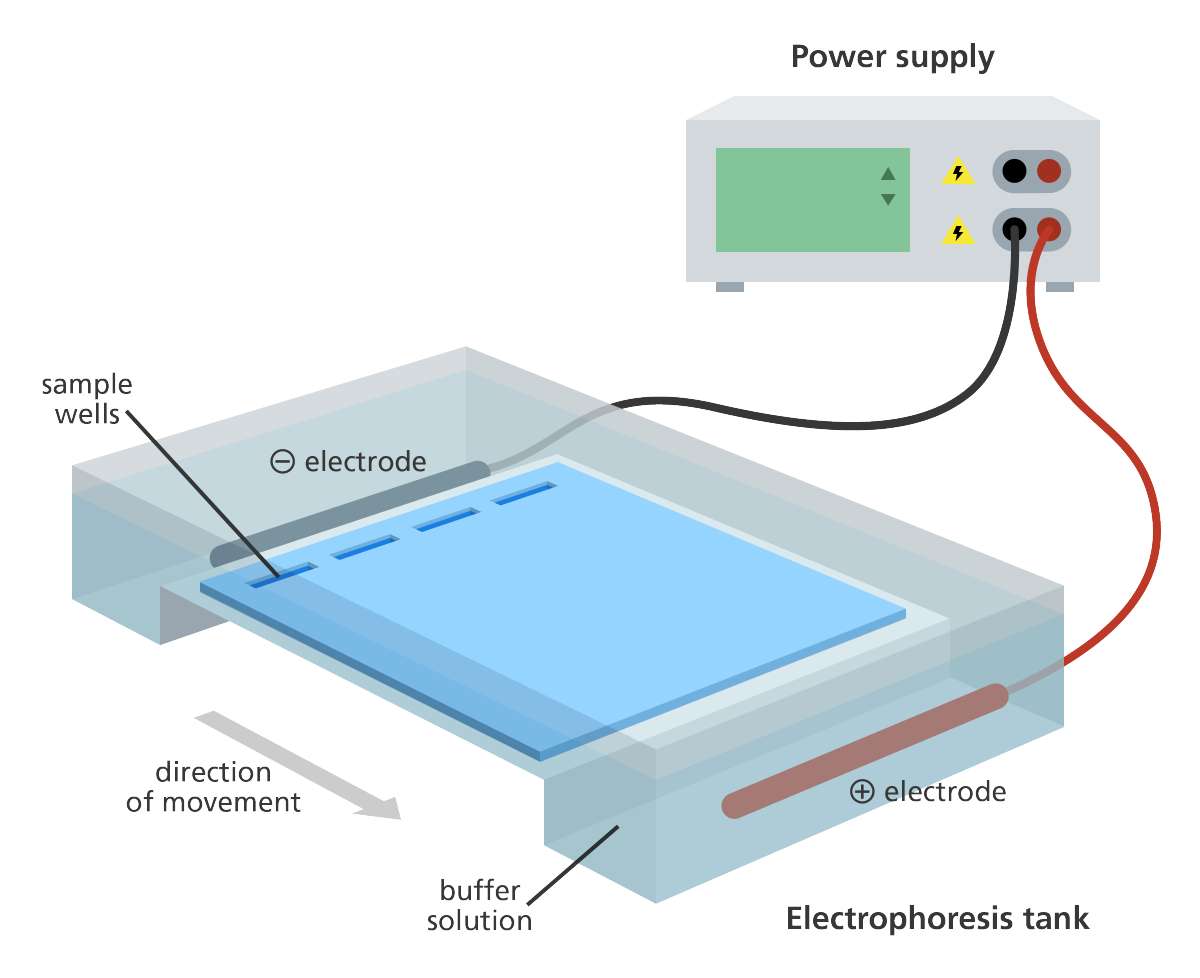
Genome Research Limited | License: CC BY 4.0
Real-time PCR (qPCR)
Measures the amount of DNA during each cycle of the reaction. Exponecial phase of the reaction is the best time to collect and analyse data.
SYBR Green I and TaqMan assays
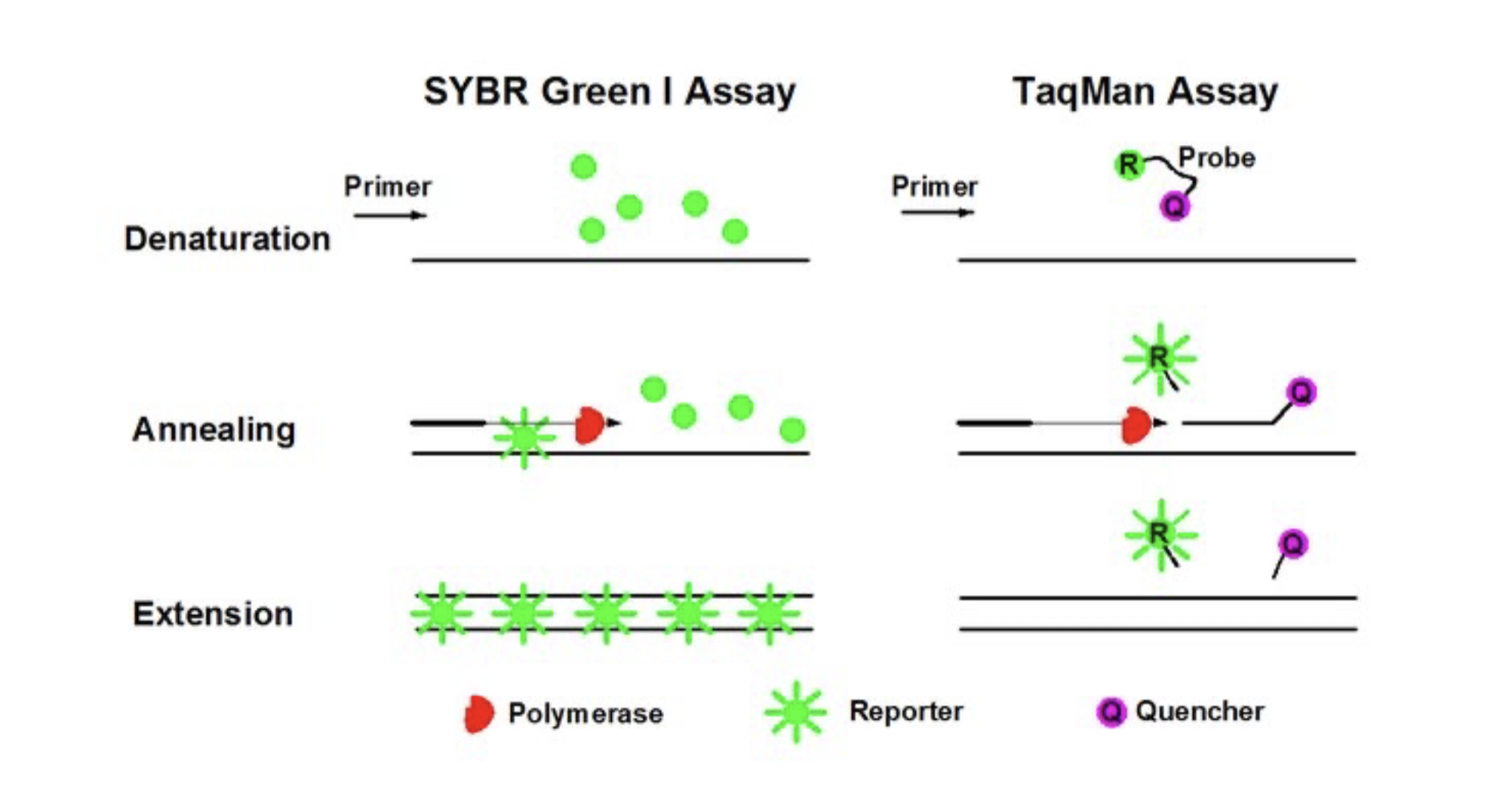
Cao, Yiqi & Luo, Yu-Feng et al. (2020). Digital PCR as an Emerging Tool for Monitoring of Microbial Biodegradation. Molecules. 25. 706. 10.3390/molecules25030706.
TaqMan Probe-Based
- TaqMan probe: a short single-stranded DNA molecule that is complementary to the target sequence.
Hybridization assay
- Hybridization: the process of forming a double-stranded nucleic acid molecule from two complementary single strands of DNA or RNA.
Why do we measure gene expression?
- gene expression is identical across all cells
- tissue-specific gene expression
- disease-specific gene expression
- gene expression changes over time
Methods for gene transcription profiling
- Microarray
- RNA-seq
- RT-PCR
RT-PCR (Reverse Transcription PCR)
- Reverse transcription: the process of synthesizing a DNA molecule from an RNA template.
Why?:
- RNA is unstable and easily degraded
- In order to have more RNA for analysis, we need to convert it to a more stable form (complementary DNA, cDNA)
Components:
- cDNA: complementary DNA
- DNA polymerase: enzyme that synthesizes DNA
- Two primers: one for the start and one for the end of the target sequence
- dNTPs: building blocks for new DNA
- Reaction buffer with Magnesium ions for enzyme activity
- Thermal cycler for temperature control
Negative control:
- No reverse transcriptase
- No template
Why examine protein expression?
- RNA is not always translated into protein
- Study correlation between RNA and protein expression
- Examine post-translational modifications
Quantification of protein expression
- Enzyme-linked immunosorbent assay (ELISA)
- Western blotting
- Immunohistochemistry (IHC)
Semi-quantitative methods: indirect measurement of expression
Quantitative methods: direct measurement of expression
Western blotting is a semi-quantitative method that uses antibodies to detect specific proteins in a sample.
Immunohistochemistry (IHC) is a semi-quantitative method that uses antibodies to detect specific proteins in a tissue section.
Immuno-staining: \
- qualitive not quantitive \
- Quantification of signal intensity and/or localisation